|
|
|
CD36 Signaling in Diabetic Cardiomyopathy
Aging and disease
2021, 12 (3):
826-840.
DOI: 10.14336/AD.2020.1217
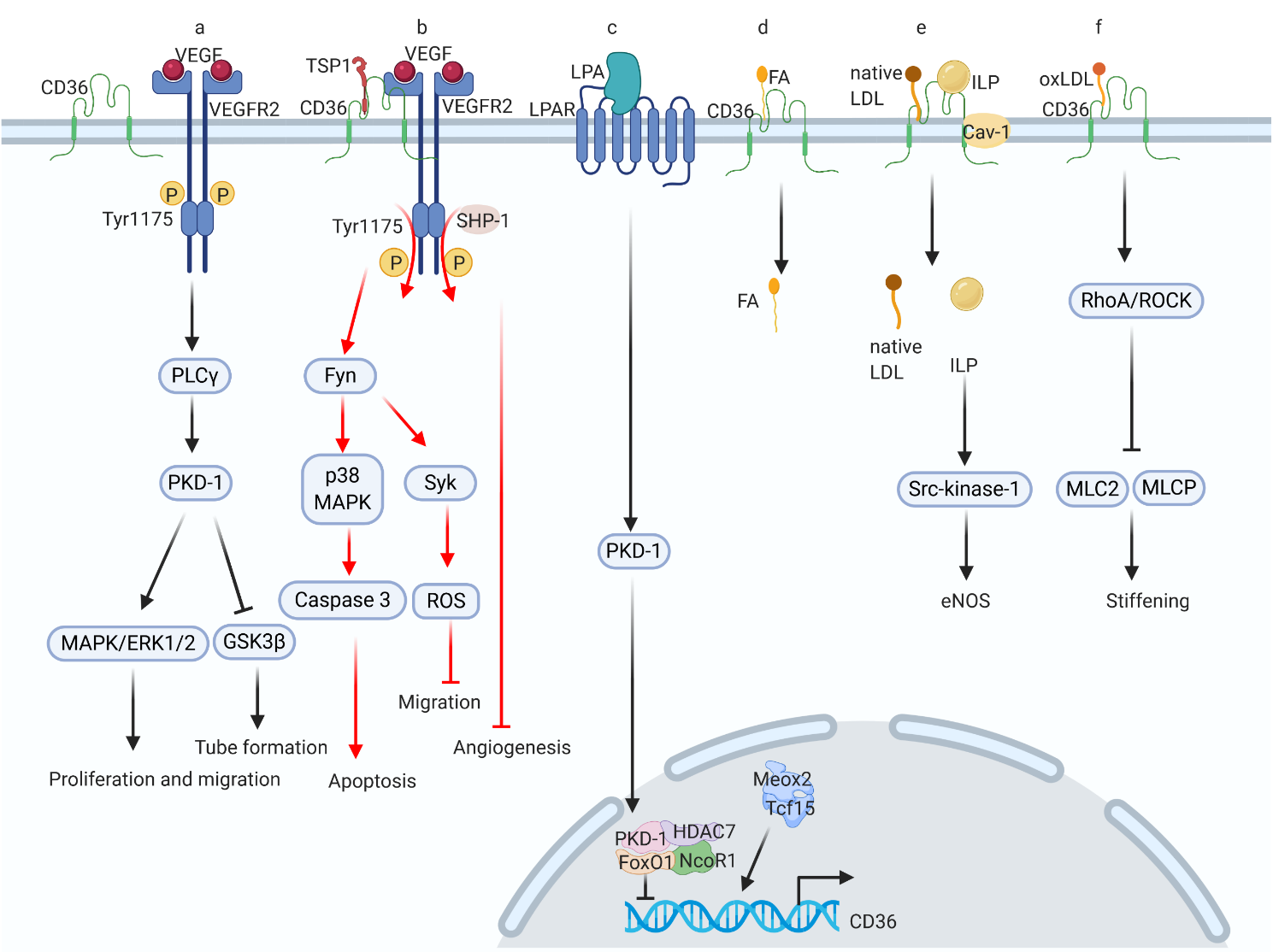
Cluster of differentiation 36 (CD36), also referred to as scavenger receptor B2, has been shown to serve multiple functions in lipid metabolism, inflammatory signaling, oxidative stress, and energy reprogramming. As a scavenger receptor, CD36 interacts with various ligands, such as oxidized low-density lipoprotein (oxLDL), thrombospondin 1 (TSP-1), and fatty acid (FA), thereby activating specific downstream signaling pathways. Cardiac CD36 is mostly expressed on the surface of cardiomyocytes and endothelial cells. The pathophysiological process of diabetic cardiomyopathy (DCM) encompasses diverse metabolic abnormalities, such as enhanced transfer of cardiac myocyte sarcolemmal FA, increased levels of advanced glycation end-products, elevation in oxidative stress, impaired insulin signaling cascade, disturbance in calcium handling, and microvascular rarefaction which are closely related to CD36 signaling. This review presents a summary of the CD36 signaling pathway that acts mainly as a long-chain FA transporter in cardiac myocytes and functions as a receptor to bind to numerous ligands in endothelial cells. Finally, we summarize the recent basic research and clinical findings regarding CD36 signaling in DCM, suggesting a promising strategy to treat this condition. 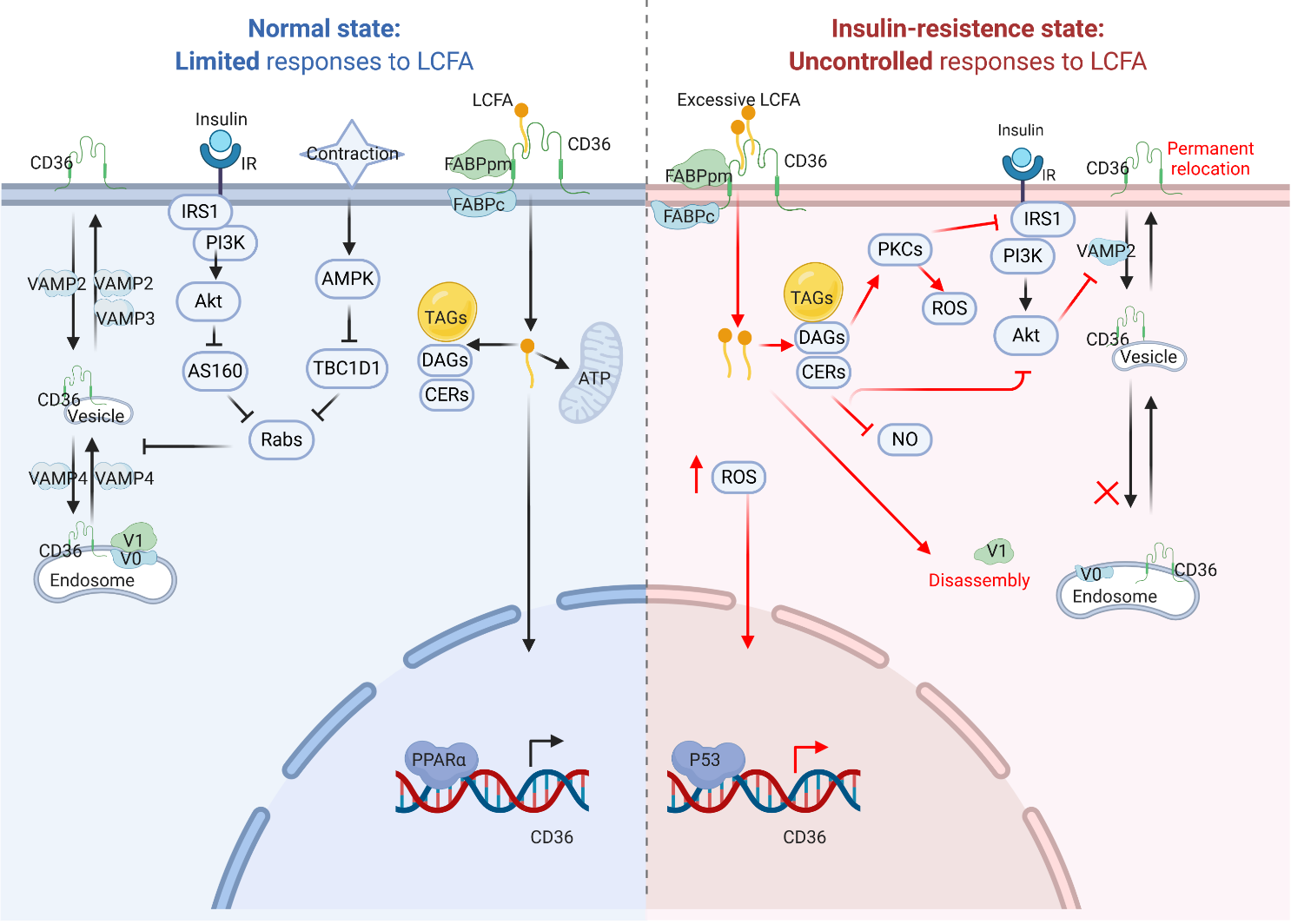
Figure 2. CD36 signaling pathway in cardiomyocytes. Under normal conditions (left), CD36 transfers LCFAs into cardiomyocytes, and LCFAs can promote CD36 transcription via PPARα binding to PPRE. Moreover, CD36 recycling occurs between endosomes and sarcolemma upon physiological stress, such as insulin stress and muscle contraction. During conditions of lipids oversupply (right), increased CD36 activity can enhance LCFA uptake to cause insulin resistance, endosome-sarcolemma recycling abnormalities and increased ROS production. LCFA, long-chain fatty acid; FABPpm, plasma membrane-localized fatty acid binding protein; FABPc, cytoplasmic FABP; TAG, triacylglycerol; DAG, diacylglycerol; Cer, ceramide; FAO, fatty acid oxidation; PPARα, peroxisome proliferator-activated receptor alpha; PPRE, PPAR response element; TSS, transcription start site; AMPK, adenosine 5’-monophosphate (AMP)-activated protein kinase; TBC1D1, Tre-2/BUB2/cdc1 domain family 1; Rabs, Rab GTPase-activating proteins; IRS1, insulin receptor substrate 1; PI3K, phosphatidylinositol 3-kinase; AKT, protein kinase B; AS160, Akt substrate 160; GLUT4, translocation of glucose transport 4; PKC, protein kinase C; V1, v-ATPase sub-complex V1; V0, v-ATPase sub-complex V0; VAMP, vesicle-associated membrane protein.
Other Images/Table from this Article
|